Research challenges:
Our overall goal is to understand how chromatin structure is employed in making cellular fate decisions, its dynamics, and how it is shaped and maintained by different chromatin regulators (CRs). We currently focus on the specific areas described below, merging basic biology, genomics and technology development.
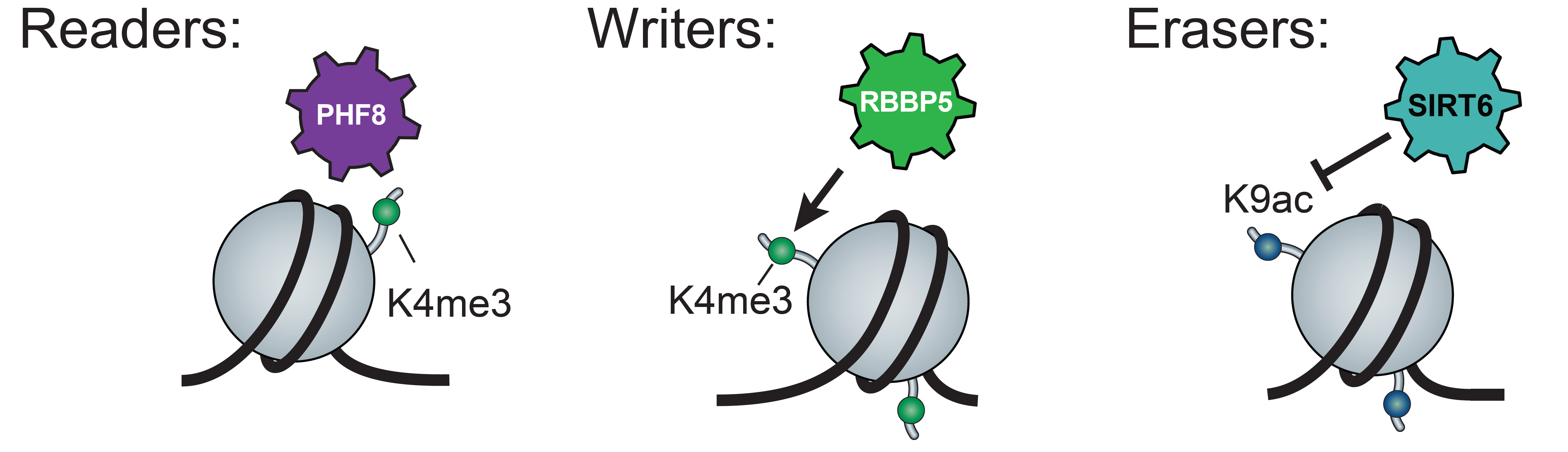
Study the principles of the genomic CR organization and recruitment
|
|
|
Chromatin structure, the genome-wide organization of DNA-associated proteins, plays a key role in cellular fate decisions. Histone modifications (HMs) are set (‘written’), removed (‘erased’) and bound (‘read’) by enzyme complexes termed Chromatin Regulators (CRs). Multiple recent studies have demonstrated that aberrations in the function of CRs are highly implicated in diseases such as cancer and developmental disorders. In our previous efforts we initiated a systematic study of the genomic organization of CRs. Following up on this, we employ our high-throughput automated ChIP-seq approach to map CRs along differentiation and use the data to computationally identify and predict key developmental CRs. We then test these predictions by perturbing the protein levels of these candidate CRs by inducible degradation and assaying the effects on differentiation.
|
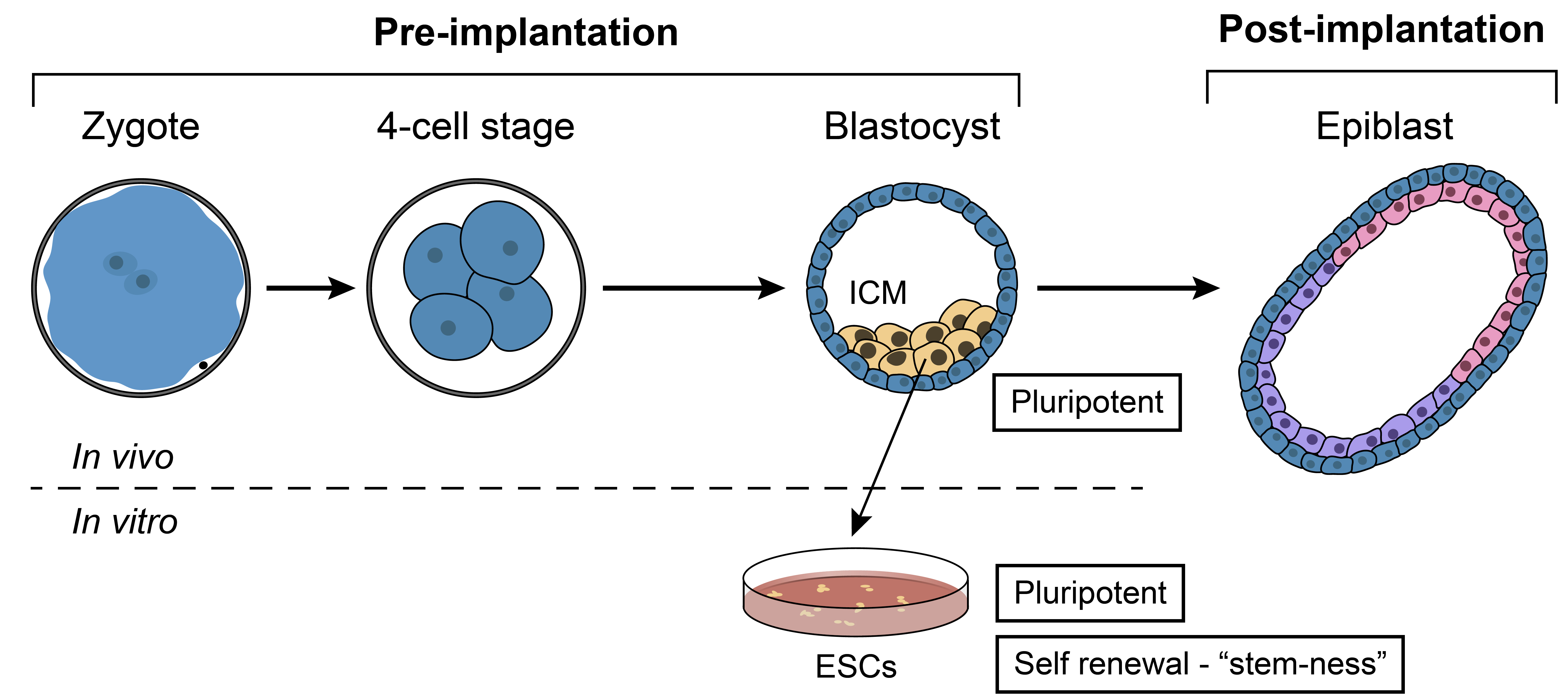
Investigating early developmental chromatin regulatory circuits in vivo and in vitro
|
|
|
In vitro portraits of embryogenesis are obtained by tissue culture of pluripotent stem cells. Yet, the faithfulness of these models is limited: for instance, while in vitro cells exhibit self-renewal, in vivo cells continue on a developmental trajectory. To reveal the regulatory circuits contributing to the distinct identity of each system, we merge quantitative immunofluorescence with genomic tools focusing on DNA associated proteins. Particularly, we employ single cell RNA-seq and high-sensitivity automated ChIP-seq to allow study of the in vivo small cell numbers, and use genetic tools to perturb candidate genes identified in our genomic analysis.
|
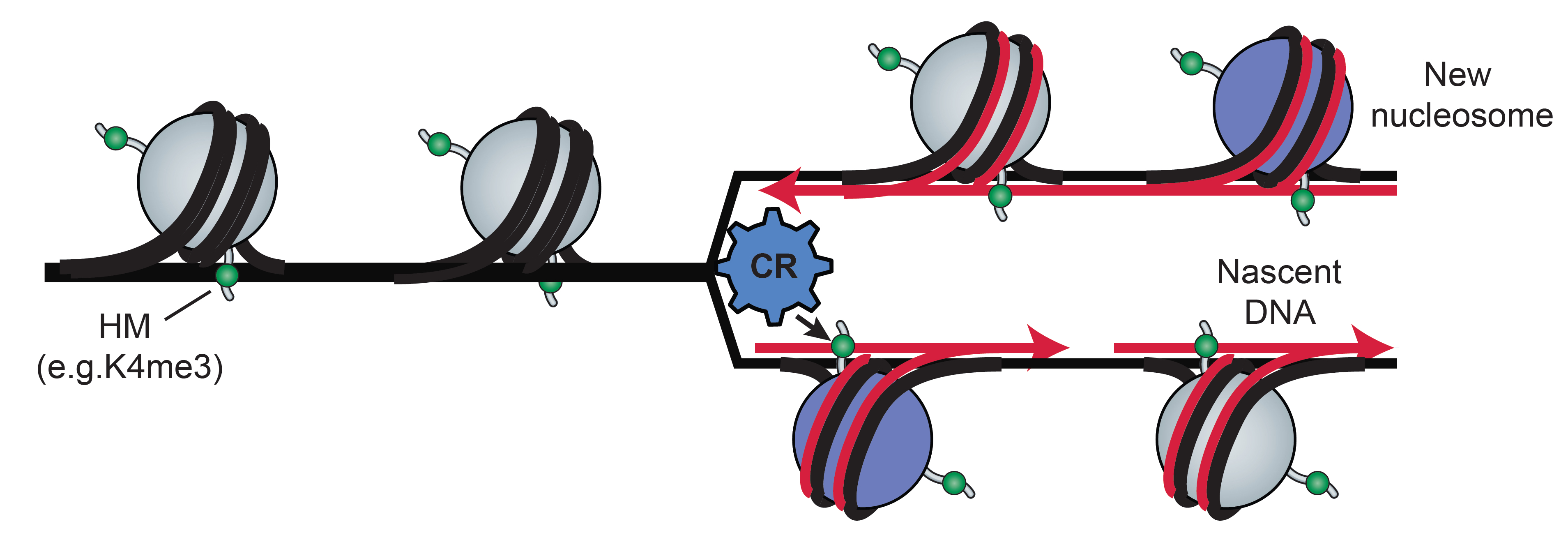
Characterize cell cycle associated chromatin dynamics and epigenetic memory
|
|
|
Both DNA sequence and the organization of DNA associated proteins are transmitted during cell divisions. This organization is disrupted during the cell cycle, and the original structure of chromatin must be restored after each cell division, a process termed ‘epigenetic memory’. This project, in collaboration with the Simon and Gymrek labs, focuses on the mechanisms that provide cellular epigenetic memory. We merge cell cycle synchronization methods, automated ChIP-seq and computational framework to model temporal dynamics of CRs and histone modifications during the cell cycle and predict critical regulators of cell cycle epigenomic maintenance. We will functionally evaluate these predictions using an inducible degradation system to perturb specific CRs and then follow the dynamics of the histone modifications in the perturbed cells.
|


